What it does: Processes single‑cell sequencing reads, partitions the genome into non‑overlapping windows, and applies Circular Binary Segmentation (CBS) together with a Hidden Markov Model (HMM) to call copy number variants (CNVs) and infer ploidy and sex.
Result(s):
- A set of CNV calls with ploidy and sex assignments that allows researchers to quickly identify chromosomal abnormalities and aneuploidies in individual samples.
- Tabular summary for each individual sample.
- Genome‑wide CNV visualization plots (per‑chromosome and whole‑genome views).
- In 2 publications (Daughtry et al., 2019; Brooks et al., 2022)
GitHub: https://github.com/melissayan/vnowchi
Click to see algorithm information
Variable Non-Overlapping Window CBS and HMM Intersect (VNOWCHI)
By counting expected versus observed single-cell sequencing reads per dynamically sized window, Circular Binary Segmentation (CBS) pinpoints change points while the Hidden Markov Model (HMM) evaluates probable state transitions. Intersecting both CBS and HMM calls (CHI) yields high‑confidence copy number variant (CNV) calls which are then classified based on rules: - filter out non‑whole genome amplification samples,
- categorize whole‑chromosome versus segmental CNVs,
- assign ploidy (euploid, aneuploid, chaotic aneuploid) and sex (female, male, unknown), and
- finally aggregate sample results to label each embryo as euploid, aneuploid, or mosaic.
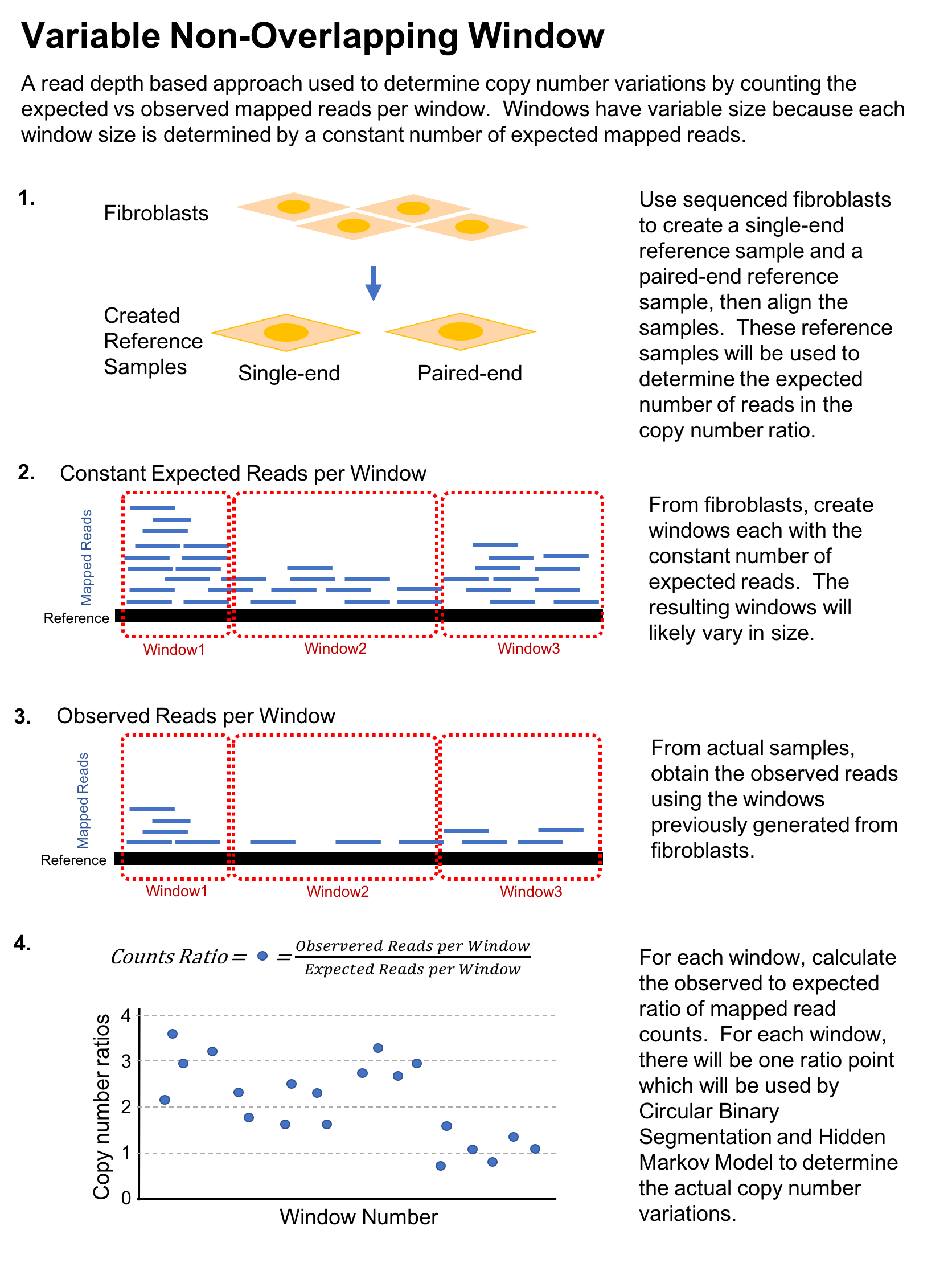
After alignment, each chromosome is divided into variable‑size, non‑overlapping windows that contain a constant expected number of mapped reads.
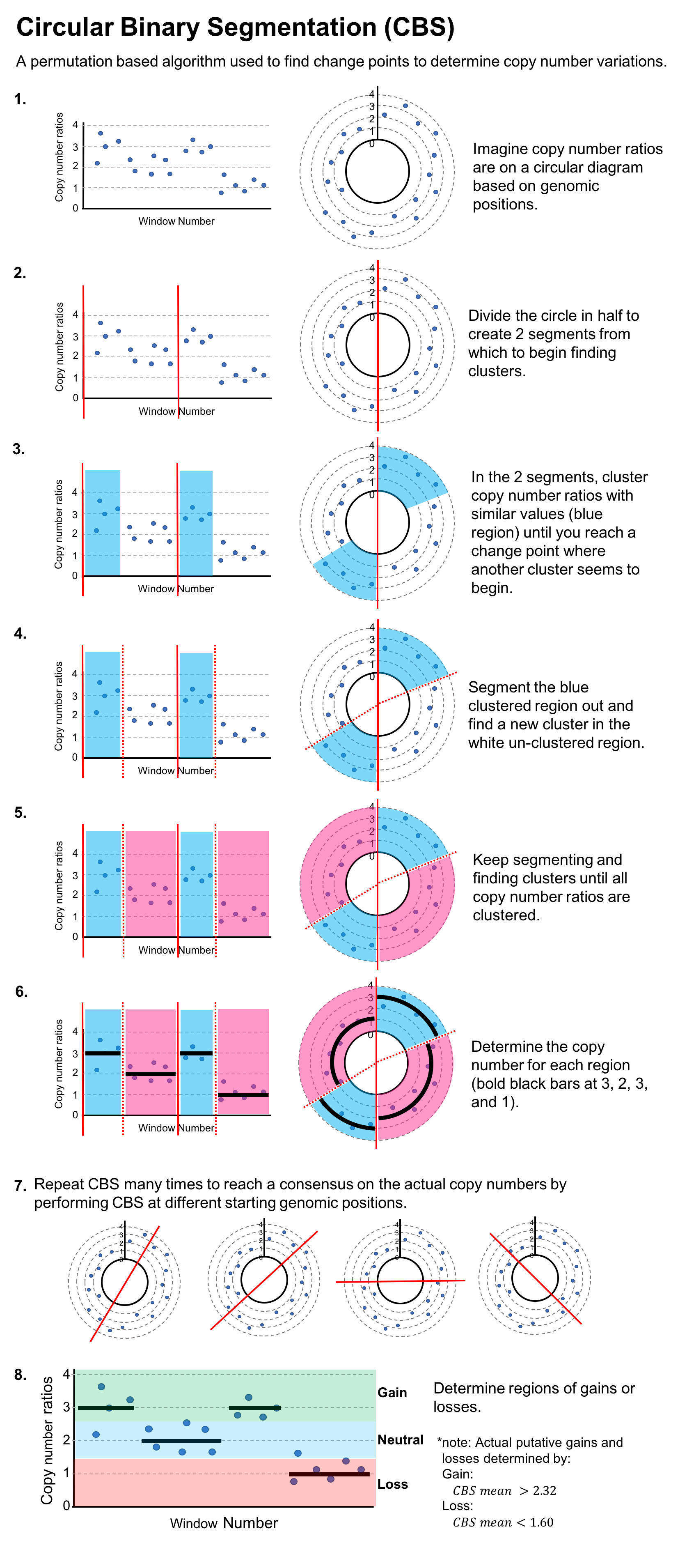
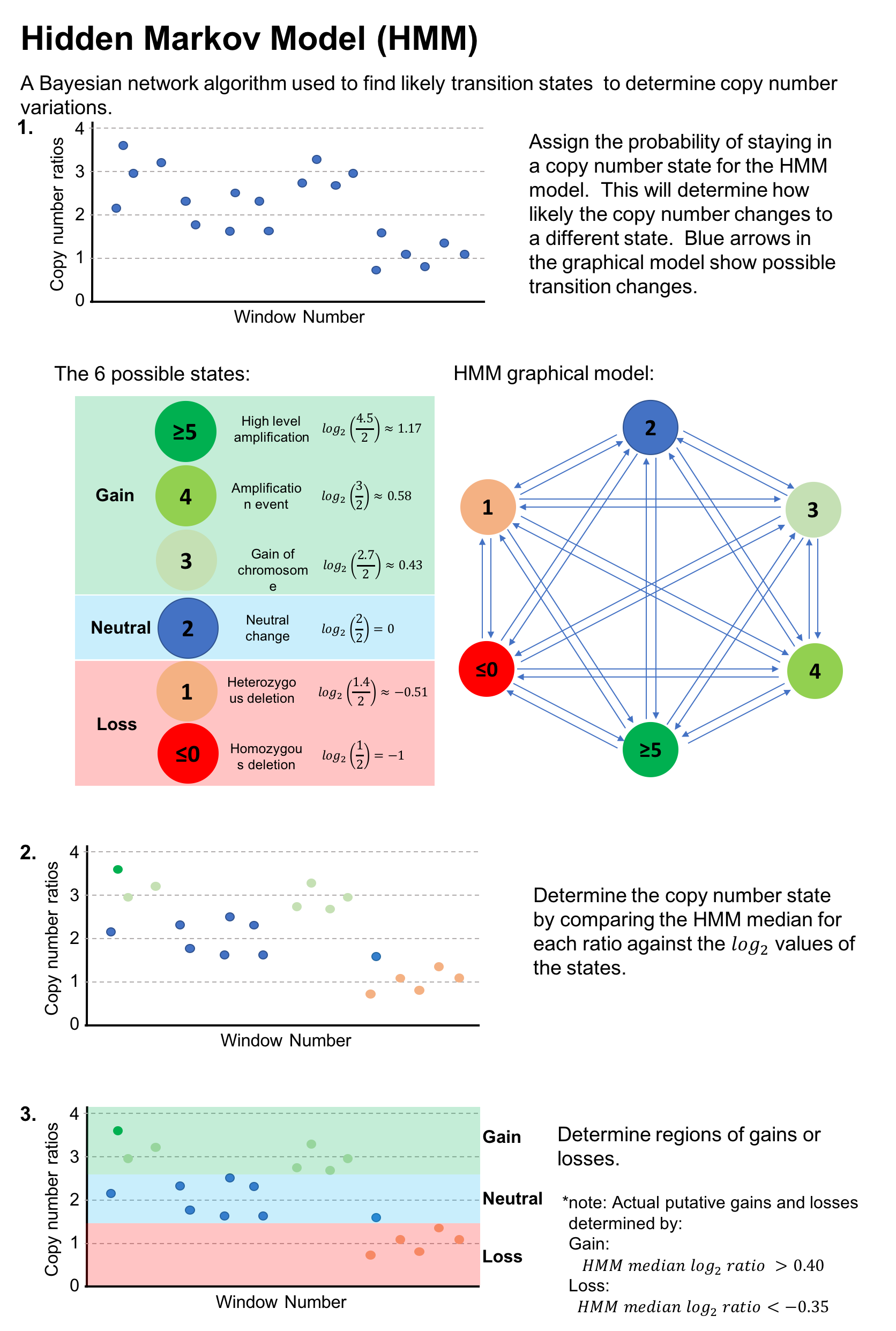
Circular Binary Segmentation (CBS) and a Hidden Markov Model (HMM) are applied to the windowed read depths to detect copy number variations.
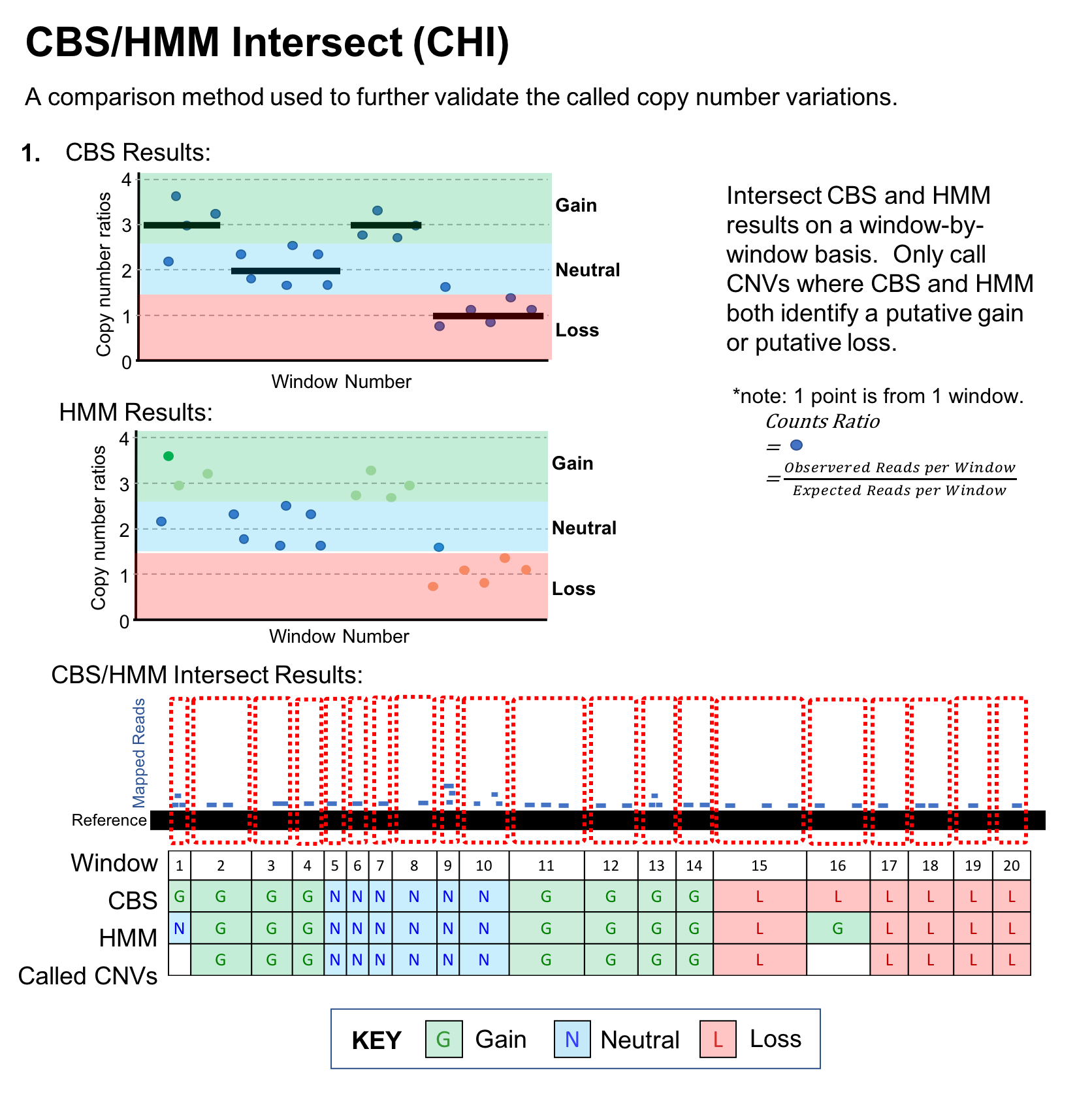
The CNV calls from CBS and HMM are intersected (CHI) to retain only high‑confidence copy number variations.
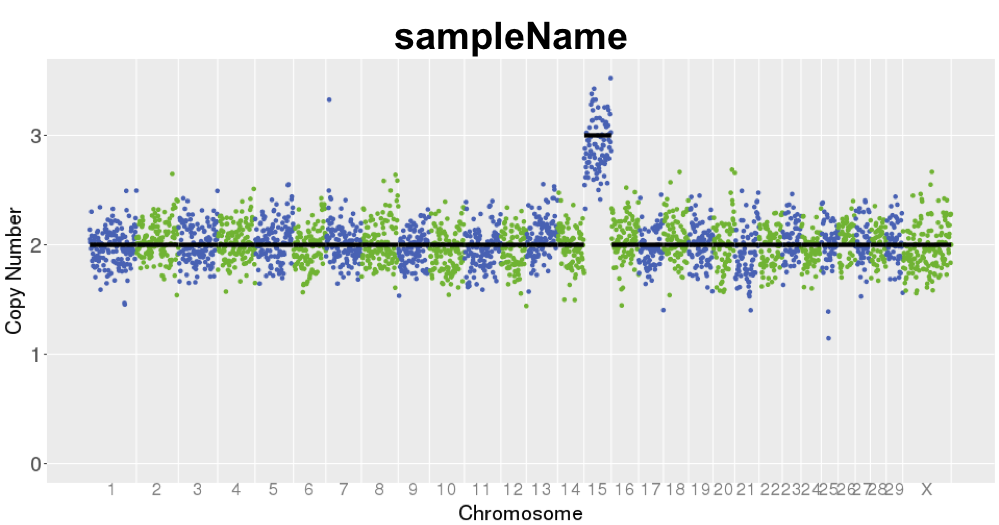
Example VNOWCHI output plot.
References
2022
-
Molecular contribution to embryonic aneuploidy and karyotypic complexity in initial cleavage divisions of mammalian development
Kelsey E Brooks, Brittany L Daughtry, Brett Davis, Melissa Y Yan, Suzanne S Fei, Selma Shepherd, Lucia Carbone, and Shawn L Chavez
Development, Apr 2022
Embryonic aneuploidy is highly complex, often leading to developmental arrest, implantation failure or spontaneous miscarriage in both natural and assisted reproduction. Despite our knowledge of mitotic mis-segregation in somatic cells, the molecular pathways regulating chromosome fidelity during the error-prone cleavage-stage of mammalian embryogenesis remain largely undefined. Using bovine embryos and live-cell fluorescent imaging, we observed frequent micro-/multi-nucleation of mis-segregated chromosomes in initial mitotic divisions that underwent unilateral inheritance, re-fused with the primary nucleus or formed a chromatin bridge with neighboring cells. A correlation between a lack of syngamy, multipolar divisions and asymmetric genome partitioning was also revealed, and single-cell DNA-seq showed propagation of primarily non-reciprocal mitotic errors. Depletion of the mitotic checkpoint protein BUB1B (also known as BUBR1) resulted in similarly abnormal nuclear structures and cell divisions, as well as chaotic aneuploidy and dysregulation of the kinase-substrate network that mediates mitotic progression, all before zygotic genome activation. This demonstrates that embryonic micronuclei sustain multiple fates, provides an explanation for blastomeres with uniparental origins, and substantiates defective checkpoints and likely other maternally derived factors as major contributors to the karyotypic complexity afflicting mammalian preimplantation development.
2019
-
Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion
Brittany L Daughtry, Jimi L Rosenkrantz, Nathan H Lazar, Suzanne S Fei, Nash Redmayne, Kristof A Torkenczy, Andrew Adey, Melissa Yan, and 5 more authors
Genome Research, Mar 2019
Aneuploidy that arises during meiosis and/or mitosis is a major contributor to early embryo loss. We previously showed that human preimplantation embryos encapsulate missegregated chromosomes into micronuclei while undergoing cellular fragmentation and that fragments can contain chromosomal material, but the source of this DNA was unknown. Here, we leveraged the use of a nonhuman primate model and single-cell DNA-sequencing (scDNA-seq) to examine the chromosomal content of 471 individual samples comprising 254 blastomeres, 42 polar bodies, and 175 cellular fragments from a large number (N = 50) of disassembled rhesus cleavage-stage embryos. Our analysis revealed that the aneuploidy and micronucleation frequency is conserved between humans and macaques, and that fragments encapsulate whole and/or partial chromosomes lost from blastomeres. Single-cell/fragment genotyping showed that these chromosome-containing cellular fragments (CCFs) can be maternally or paternally derived and display double-stranded DNA breaks. DNA breakage was further indicated by reciprocal subchromosomal losses/gains between blastomeres and large segmental errors primarily detected at the terminal ends of chromosomes. By combining time-lapse imaging with scDNA-seq, we determined that multipolar divisions at the zygote or two-cell stage were associated with CCFs and generated a random mixture of chromosomally normal and abnormal blastomeres with uniparental or biparental origins. Despite frequent chromosome missegregation at the cleavage-stage, we show that CCFs and nondividing aneuploid blastomeres showing extensive DNA damage are prevented from incorporation into blastocysts. These findings suggest that embryos respond to chromosomal errors by encapsulation into micronuclei, elimination via cellular fragmentation, and selection against highly aneuploid blastomeres to overcome chromosome instability during preimplantation development.